| dyads_test_vs_ctrl_m1_shift0 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=13; shift=0; ncol=13; wmGGACGCGGTkk
; Alignment reference
a 31 38 15 8 104 18 9 10 6 7 12 26 21
c 21 29 9 10 1 82 5 90 3 7 9 20 19
g 27 18 78 87 5 6 93 6 100 85 6 34 31
t 34 28 11 8 3 7 6 7 4 14 86 33 42
|
| CMTA2_MA0969.1_JASPAR_rc_shift4 (CMTA2:MA0969.1:JASPAR_rc) |
 |
0.906 |
0.627 |
0.749 |
0.940 |
0.968 |
1 |
2 |
3 |
1 |
1 |
1.600 |
1 |
; dyads_test_vs_ctrl_m1 versus CMTA2_MA0969.1_JASPAR_rc (CMTA2:MA0969.1:JASPAR_rc); m=1/5; ncol2=9; w=9; offset=4; strand=R; shift=4; score= 1.6; ----ACRCGstww
; cor=0.906; Ncor=0.627; logoDP=0.749; NsEucl=0.940; NSW=0.968; rcor=1; rNcor=2; rlogoDP=3; rNsEucl=1; rNSW=1; rank_mean=1.600; match_rank=1
a 0 0 0 0 928 15 280 1 11 36 243 260 264
c 0 0 0 0 54 970 7 970 8 306 243 176 220
g 0 0 0 0 11 7 682 11 955 438 121 237 203
t 0 0 0 0 7 8 31 18 26 220 393 327 313
|
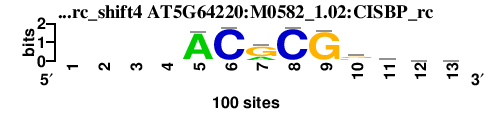
| AT5G64220_M0582_1.02_CISBP_rc_shift4 (AT5G64220:M0582_1.02:CISBP_rc) |
 |
0.905 |
0.627 |
0.755 |
0.940 |
0.968 |
2 |
3 |
2 |
2 |
2 |
2.200 |
2 |
; dyads_test_vs_ctrl_m1 versus AT5G64220_M0582_1.02_CISBP_rc (AT5G64220:M0582_1.02:CISBP_rc); m=2/5; ncol2=9; w=9; offset=4; strand=R; shift=4; score= 2.2; ----ACRCGsyww
; cor=0.905; Ncor=0.627; logoDP=0.755; NsEucl=0.940; NSW=0.968; rcor=2; rNcor=3; rlogoDP=2; rNsEucl=2; rNSW=2; rank_mean=2.200; match_rank=2
a 0 0 0 0 93 2 28 0 1 3 24 25 26
c 0 0 0 0 5 97 1 97 1 31 25 18 22
g 0 0 0 0 1 1 68 1 95 44 12 24 20
t 0 0 0 0 1 0 3 2 3 22 39 33 32
|
| CAMTA1_MA1197.1_JASPAR_rc_shift1 (CAMTA1:MA1197.1:JASPAR_rc) |
 |
0.729 |
0.673 |
3.455 |
0.911 |
0.904 |
5 |
1 |
1 |
5 |
5 |
3.400 |
3 |
; dyads_test_vs_ctrl_m1 versus CAMTA1_MA1197.1_JASPAR_rc (CAMTA1:MA1197.1:JASPAR_rc); m=3/5; ncol2=12; w=12; offset=1; strand=R; shift=1; score= 3.4; -yhmACGCGytTt
; cor=0.729; Ncor=0.673; logoDP=3.455; NsEucl=0.911; NSW=0.904; rcor=5; rNcor=1; rlogoDP=1; rNsEucl=5; rNSW=5; rank_mean=3.400; match_rank=3
a 0 133 174 151 595 0 20 0 0 1 55 77 124
c 0 170 156 326 3 598 0 598 0 285 132 45 117
g 0 58 33 58 0 0 578 0 598 140 6 46 56
t 0 237 235 63 0 0 0 0 0 172 405 430 301
|
| CAMTA3_M0581_1.02_CISBP_rc_shift1 (CAMTA3:M0581_1.02:CISBP_rc) |
 |
0.861 |
0.596 |
0.713 |
0.926 |
0.950 |
4 |
5 |
5 |
4 |
4 |
4.400 |
5 |
; dyads_test_vs_ctrl_m1 versus CAMTA3_M0581_1.02_CISBP_rc (CAMTA3:M0581_1.02:CISBP_rc); m=5/5; ncol2=9; w=9; offset=1; strand=R; shift=1; score= 4.4; -vbrACrCGg---
; cor=0.861; Ncor=0.596; logoDP=0.713; NsEucl=0.926; NSW=0.950; rcor=4; rNcor=5; rlogoDP=5; rNsEucl=4; rNSW=4; rank_mean=4.400; match_rank=5
a 0 25 21 28 100 0 27 1 1 4 0 0 0
c 0 25 25 21 0 96 0 96 0 18 0 0 0
g 0 26 25 27 0 0 66 0 97 54 0 0 0
t 0 24 29 24 0 4 7 3 2 24 0 0 0
|




